library(rethinking)
library(dagitty)
data(Howell1)Lecture 03 - Geocentric Models
Rose / Thorn
Rose: Non-independent variables in a summary table
Thorn: Stupid resolution to my plot coding issue
Linear Regressions
- essentially a geocentric model - overly simplified
- separate from a causal model
- associations are from the causal model, not the statistical model
Gaussian Distributions
- there are many more ways to end up in the center than to end up on the periphery
- why the Gaussian distribution spontaneously occurs in natural systems
- generative: if we add fluctuations, we tend towards normal distribution (lots of summed fluctuations in nature)
- inferential: estimating mean and variance, normal distribution is best one to use because it is least informative (no other information present)
- normal distribution is just a tool for estimating mean/variance because it has widest distribution
- data doesn’t have to be normal to be able to leverage the tool to estimate mean/variance
Workflow
- State a clear question
- Sketch causal assumptions
- Define generative model from sketch
- Use generative model to build estimator
- Profit
Describing Models
= is deterministic
- ~ is distributional
Howell Example
- Question/Estimand: Describe association between adult weight and height
d2 <- Howell1[Howell1$age>=18,]- Scientific model: weight is some function of height and unobserved influences
d <- dagitty("dag {
H -> W
U -> W
}")
drawdag(d)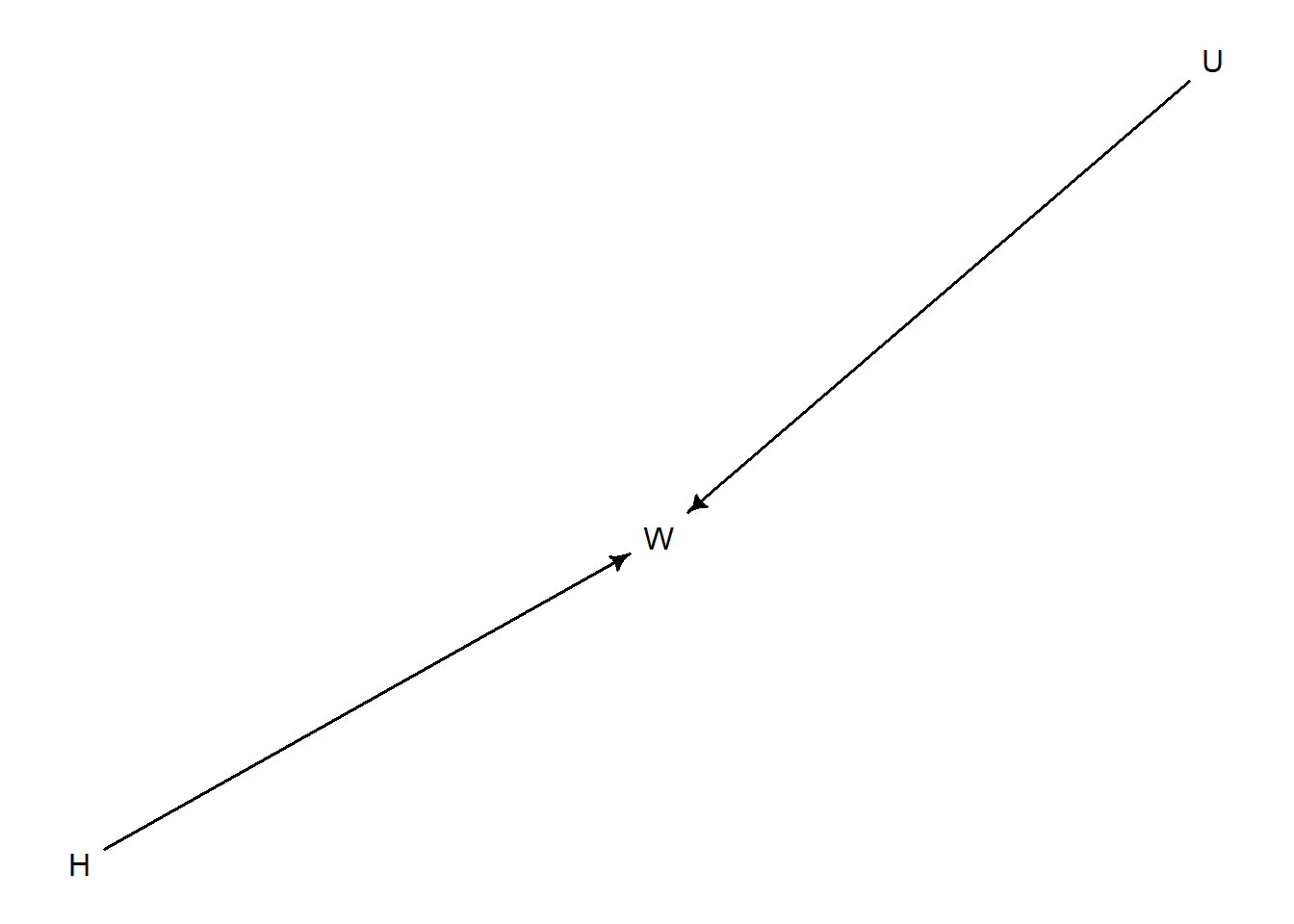
\[ W = f(H, U) \]
- Generative/Statistical model
Two options: dynamic (complex and ongoing) and static
- Static model allows us to imagine changes at specific times and still use a Gaussian distribution
For adults, weight is a proportion of height plus the influence of unobserved causes
\[ W = \beta H+U \]
sim_weight <- function(H, b, sd){
U <- rnorm(length(H), 0, sd)
W <- b*H + U
return(W)
}
H <- runif(200, min = 130, max = 170)
W <- sim_weight(H, b = 0.5, sd = 5)
plot(W ~ H, col = 2, lwd = 3)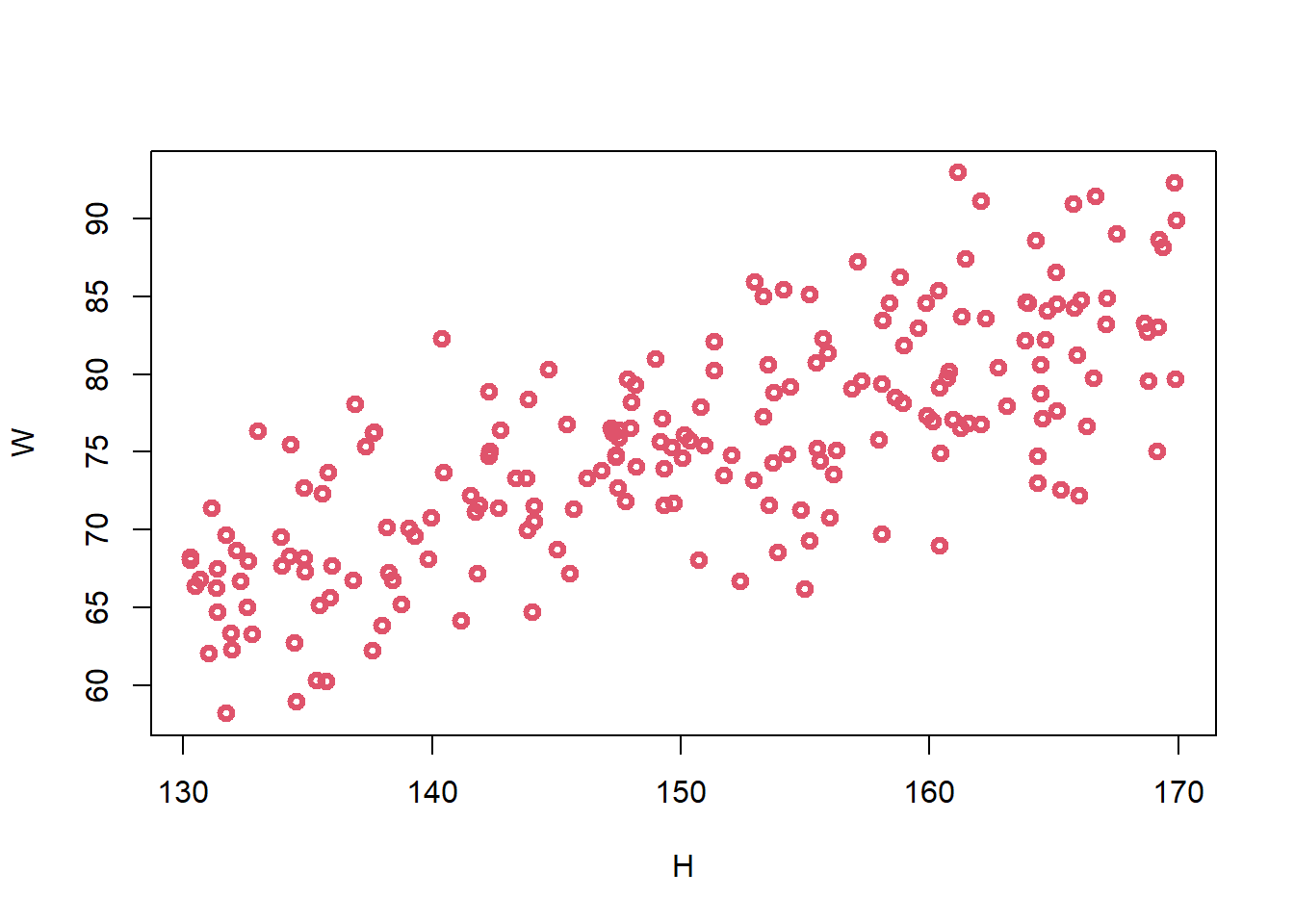
- can adjust to produce biologically unrealistic relationships by adjusting b to be large or small
\[ W_i = \beta H_i + U_i \\ U_i \sim Normal(0, \sigma) \\ H_i \sim Uniform(130, 170) \]
~ indicates distribution
= is deterministic
we want to estimate how the average weight changes with height
\[ E(W_{i}|H_{i}) = \alpha + \beta H_{i} \]
\(E(W_{i}|H_{i})\) = average weight conditional on height
\(\alpha\) = intercept (when height is 0, what is weight? Scientifically should be zero but putting it in model to make sure our model is scientifically sound)
\(\beta\) = slope
Posterior distribution:
\[ Pr(\alpha,\beta,\sigma| H_{i}, W_{i}) = \frac{Pr(W_{i}|H_{i}, \alpha,\beta,\sigma)Pr(\alpha,\beta,\sigma)}{Z} \]
alpha, beta, sigma are our three unknowns: alpha and beta define the line and sigma defines the error around it
unknowns are conditional (|) on the data Hi and Wi
alpha, sigma, beta are unknown so we need a posterior distribution for them (and they are dependent on the data)
\(Pr(\alpha,\beta,\sigma| H_{i}, W_{i})\) = posterior probability of specific line
\(Pr(W_{i}|H_{i}, \alpha,\beta,\sigma)\) = probability of each weight dependent on height value, alpha, beta, sigma (garden of forking data)
\(Pr(\alpha,\beta,\sigma)\) = prior
Z = normalizing constant
\(W_{i} \sim Normal(\mu _{i}), \sigma)\): linear model
\(\mu _{i} = \alpha + \beta H_{i}\)
Quadratic approximation (continuous version of grid approximation)
approximate posterior distribution as a multivariate Gaussian distrubution
posteriors are often Gaussian
\(W_{i} \sim Normal(\mu _{i}), \sigma)\)
\(\mu _{i} = \alpha + \beta H_{i}\)
\(\alpha \sim Normal(0,10)\)
\(\beta \sim Uniform(0,1)\)
\(\sigma \sim Uniform(0, 10)\)
H <- runif(10, 130, 170)
W <- sim_weight(H, b = 0.5, sd = 5)
m3.1 <- quap(alist(
W ~ dnorm(mu, sigma),
mu <- a + b*H,
a ~ dnorm(0, 10),
b ~ dunif(0, 1),
sigma ~ dunif(0, 10)
), data = list(W=W, H=H))Priors
- we want to constrain to scientifically plausible values
- justify with information outside the data - like the rest of model
- priors have no essential differences from posteriors except for data – we can use them as generative models and ensure that our assumptions are reasonable
n <- 1e3
a <- rnorm(n, 0, 10)
b <- runif(n, 0, 1)
plot(NULL, xlim = c(130, 170), ylim = c(50, 90)) +
for (j in 1:50) abline(a=a[j], b=b[j], col = 'red') 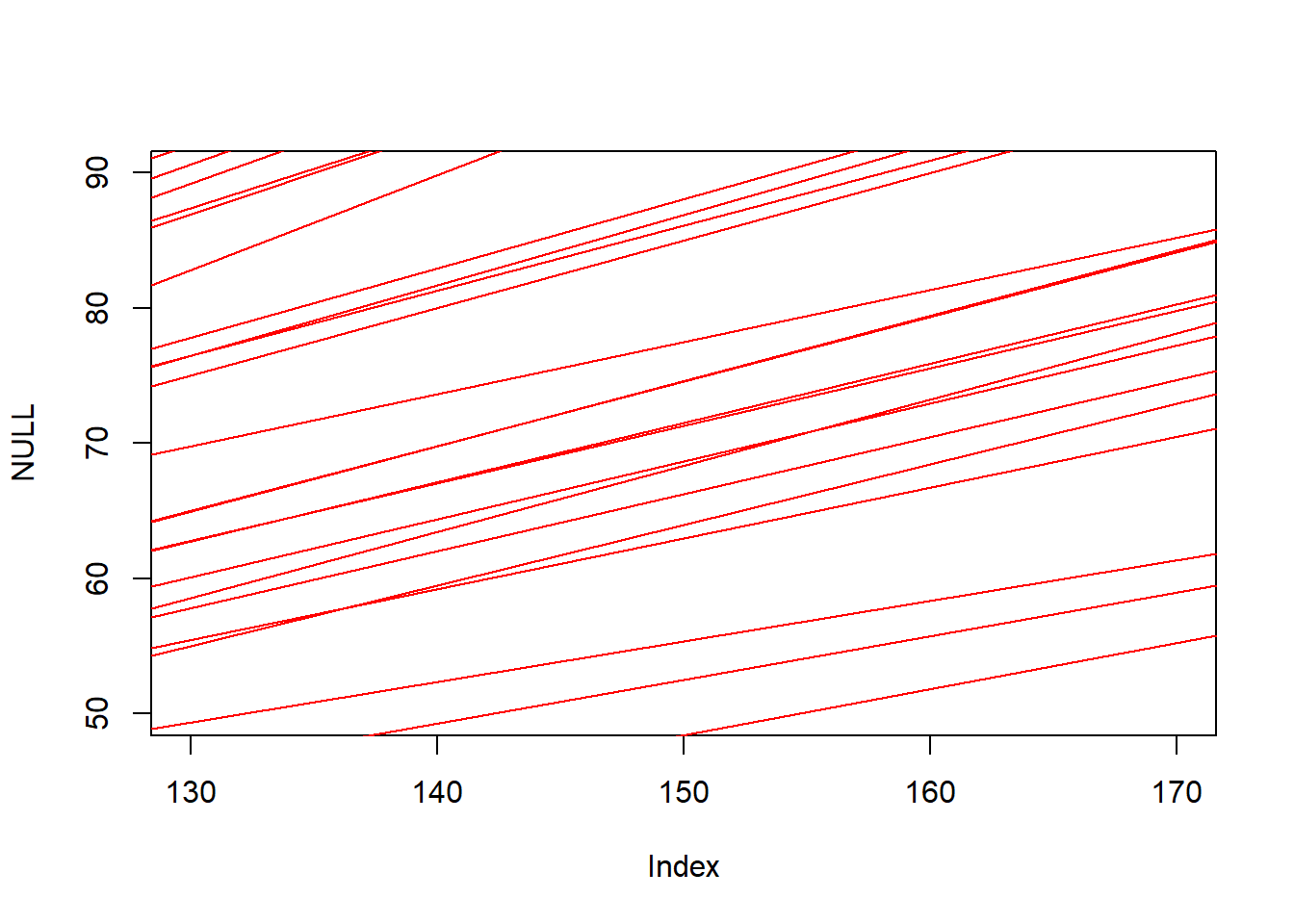
integer(0)- Validate model
test statistical model with simulated observations from scientific model - need to test if your model is working
use many values and make sure your model responds appropriately
# model summary
precis(m3.1) mean sd 5.5% 94.5%
a 0.6093014 9.50424778 -14.5803222 15.7989250
b 0.5045905 0.06551189 0.3998898 0.6092911
sigma 6.1344656 1.35444780 3.9697964 8.2991347- Analyze data
dat <- list(W = d2$weight, H = d2$height)
m3.2 <- quap(alist(
W ~ dnorm(mu, sigma),
mu <- a + b*H,
a ~ dnorm(0, 10),
b ~ dunif(0, 1),
sigma ~ dunif(0, 10)
), data = dat)
precis(m3.2) mean sd 5.5% 94.5%
a -43.7106844 4.17274593 -50.379538 -37.0418305
b 0.5738951 0.02696368 0.530802 0.6169883
sigma 4.2584569 0.16227623 3.999108 4.5178056parameters are not independent of one another, they cannot be independently interpreted
- use posterior predictions and describe/interpret those by sampling the posterior distribution
Posterior Predictive Distribution
post <- extract.samples(m3.2)
plot(d2$height, d2$weight) +
for (j in 1:20) abline(a=post$a[j], b=post$b[j])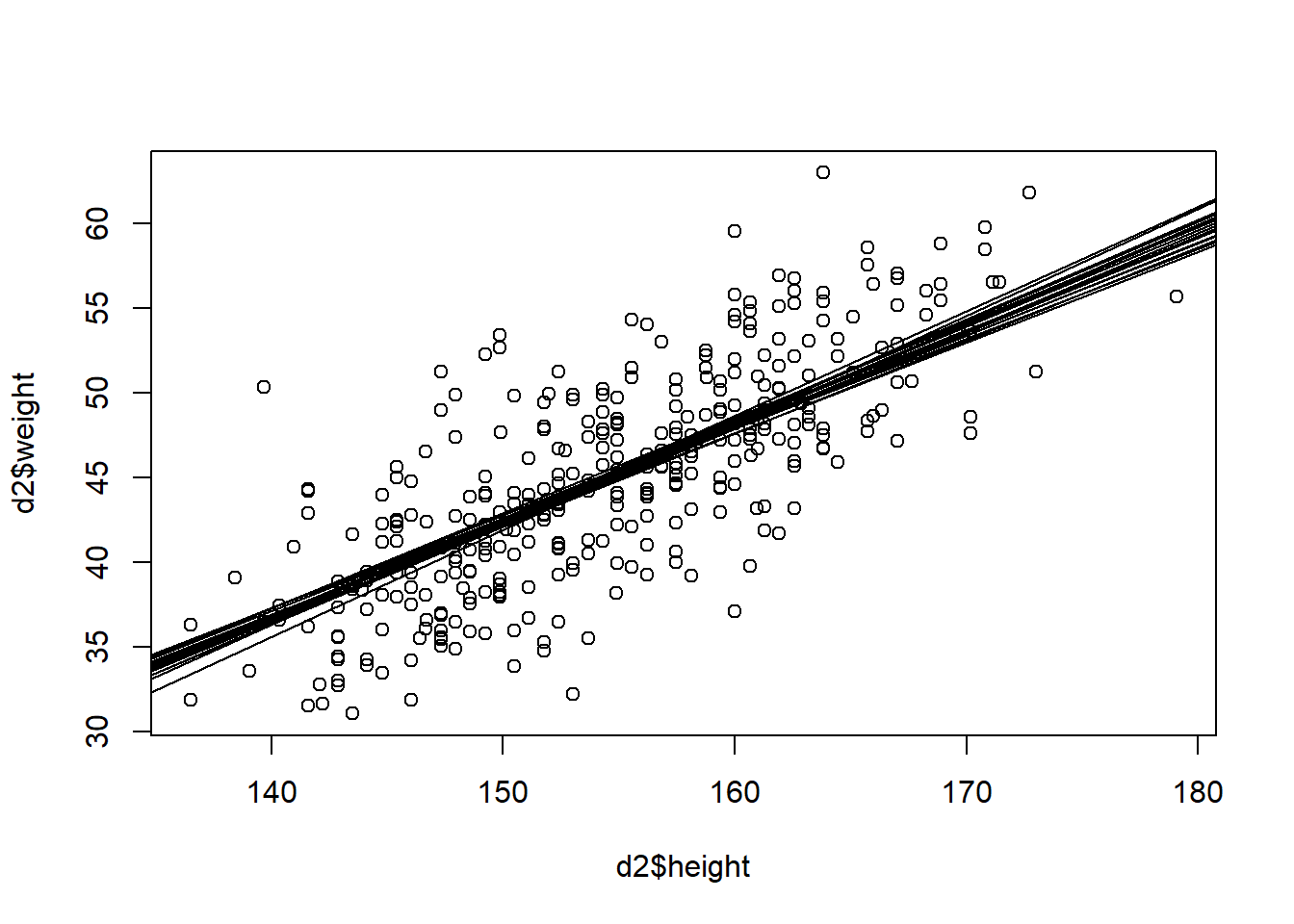
integer(0)height_seq <- seq(130, 190, len = 20)
W_postpred <- sim(m3.2, data = list(H=height_seq))
W_PI <- apply(W_postpred, 2, PI)
plot(d2$height, d2$weight) +
for (j in 1:20) abline(a=post$a[j], b=post$b[j]) +
lines(height_seq, W_PI[1,], lty = 2) +
lines(height_seq, W_PI[2,], lty = 2)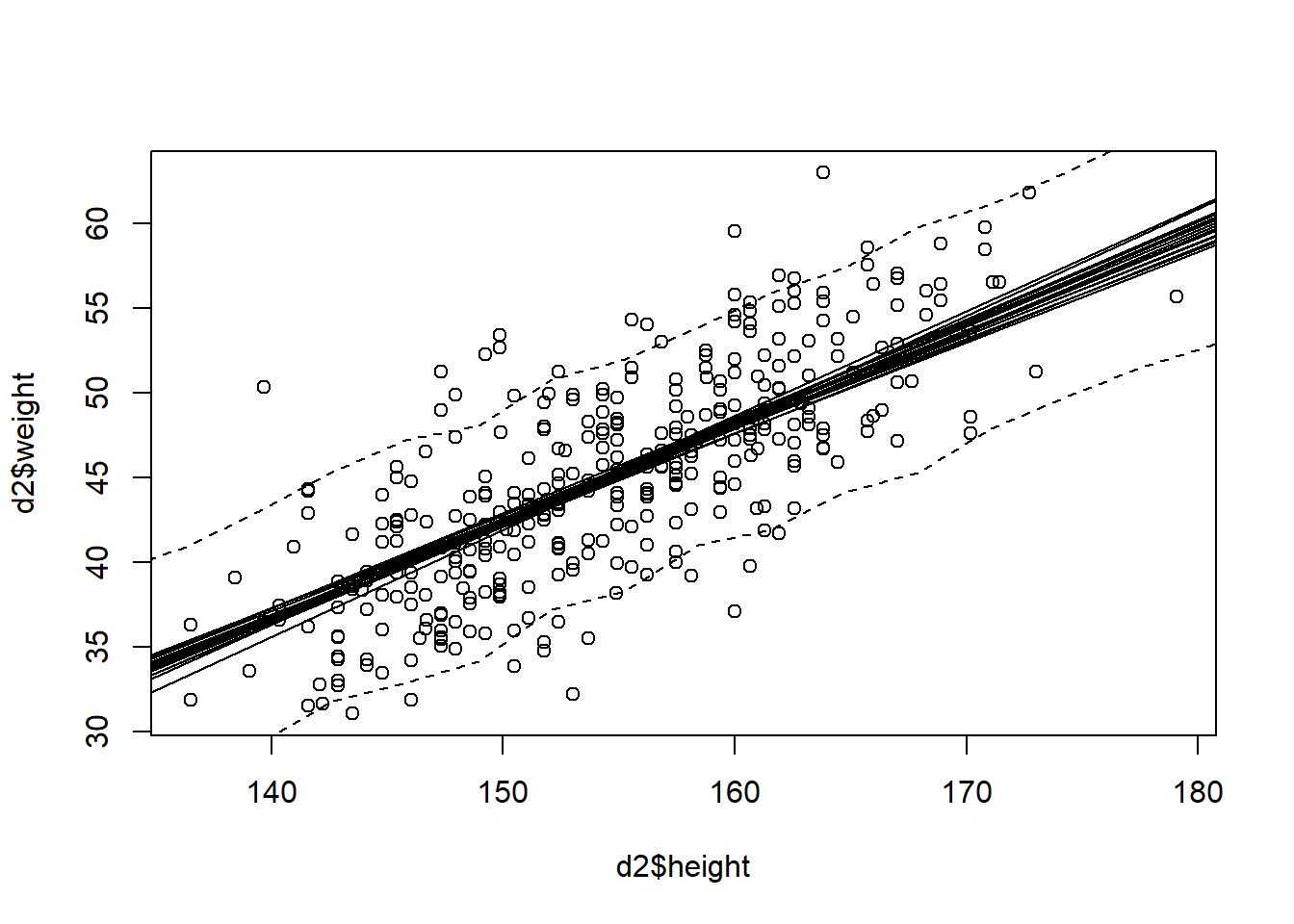
integer(0)