Lecture 08 - Markov chain Monte Carlo
Rose / Thorn
Rose:
Thorn:
Modelling Approaches
quadratic approximation makes strong assumptions about what the model looks like - approximately Gaussian
MCMC is intensive but with less assumptions and more flexible
Markov chain Monte Carlo
chain: sequence of draws from distribution
Markov chain: history doesn’t matter, just where you are now
Monte Carlo: random simulation
Hamiltonian Monte Carlo
- probability space is essentially a skate park
- uses random pathways that follow the distribution of the variables
dag <- dagify(
S ~ Q + J + X,
Q ~ X,
J ~ Z,
outcome = 'S',
latent = 'Q',
coords = list(x = c(Q = 0, X = 0, S = 1, J = 1, Z = 2),
y = c(X = 0, J = 0, Z = 0, Q = 1, S = 1))
)
ggdag(dag) + theme_dag()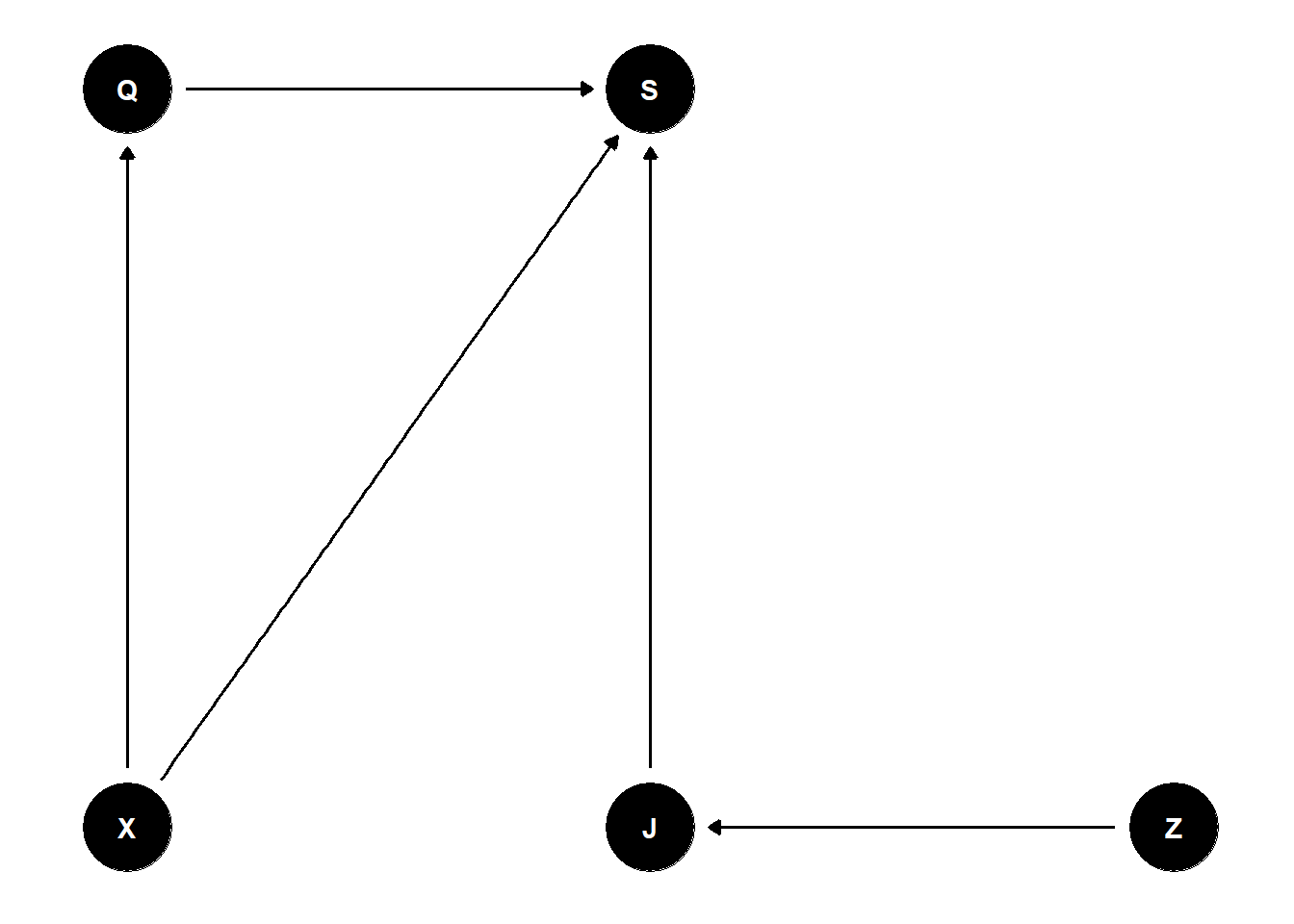
- Estimand: association between wine quality and wine origin. Stratify by judge for efficiency.
data(Wines2012)
d <- Wines2012
dat <- list(
S = standardize(d$score),
J = as.numeric(d$judge),
W = as.numeric(d$wine),
X = ifelse(d$wine.amer == 1,1,2),
Z = ifelse(d$judge.amer == 1,1,2)
)
mQ <- ulam(alist(
S ~ dnorm(mu, sigma),
mu <- Q[W],
Q[W] ~ dnorm(0, 1),
sigma ~ dexp(1)),
data = dat, chains = 4, cores = 4)
precis(mQ, 2)trace plots: visualization of the Markov chain
want more than one chain in order to check convergence
- convergence: each chain explores the right distribution and the same distribution
R-hat is chain convergence diagnostic
variance ratio
if chains do converge, beginning of the chain and end of chain should be exploring the same place and therefore the chain is stationary
as total variance (among chains) approaches average variance within chains, R-hat approaches 1
if chains were exploring different regions, the total variance would be bigger and Rhat is larger than 1
does not guarantee convergence but gives an idea that the chains are working when ~ 1
n_eff = effective samples
approximation of how long the chain would be if each sample was completely independent of the one before it (perfectly uncorrelated, truly random)
when samples are autocorrelated, you have fewer effective samples
typically n_eff is smaller than number of samples you actually took
- because perfect chain will need fewer samples/be shorter
mQO <- ulam(alist(
S ~ dnorm(mu, sigma),
mu <- Q[W] + O[X],
Q[W] ~ dnorm(0, 1),
O[X] ~ dnorm(0, 1),
sigma ~ dexp(1)),
data = dat, chains = 4, cores = 4)
plot(precis(mQO, 2))
mQOJ <- ulam(alist(
S ~ dnorm(mu, sigma),
mu <- (Q[W] + O[X] - H[J])*D[J],
Q[W] ~ dnorm(0, 1),
O[X] ~ dnorm(0, 1),
H[J] ~ dnorm(0, 1),
D[J] ~ dexp(1),
sigma ~ dexp(1)),
data = dat, chains = 4, cores = 4)
plot(precis(mQOJ, 2))adding judges adds information to the model and allows for better estimates
often if there is an issue, it is with your model
loop back to basic scientific questions and assumptions
divergent transitions are one of the signs of this - “rejected proposal”